
GSL Biotech's software for planning and visualization in molecular biology genetic research.SnapGene"ofLatest version 6.0 Was officially released (November 2021, 11 local time).
This major update includes restriction enzyme editing with silent mutations, an expanded set of feature types, shareable agarose gel files, and enhanced cloning simulation capabilities for more flexible editing and browsing. became.
This article references What's New in SnapGene?
https://go.snapgene.com/whats-new
table of contents
About SnapGene

This is software for planning, visualizing, and documenting genetic analysis using molecular biological methods that supports cloning and PCR operations. Document the DNA constructs you create in your research in an easy-to-read electronic format. These document files can be shared among researchers using a free viewer (SnapGene Viewer).
Main functions

- Molecular Cloning Techniques
– Simulation of PCR and common cloning methods
– Visualization of DNA, chromosomes (chromosome size sequences), etc.
– Automatically records all operations during the cloning procedure
- Align & Verify
– Error detection for accurate cloning
– Plasmid sequence and annotation, alignment, agarose gel electrophoresis - Data management
– Automatically create a change history with excellent identification such as color coding
– Supports various file formats
– Convert various file formats in molecular biology to SnapGene
Version 6.0 What's new in
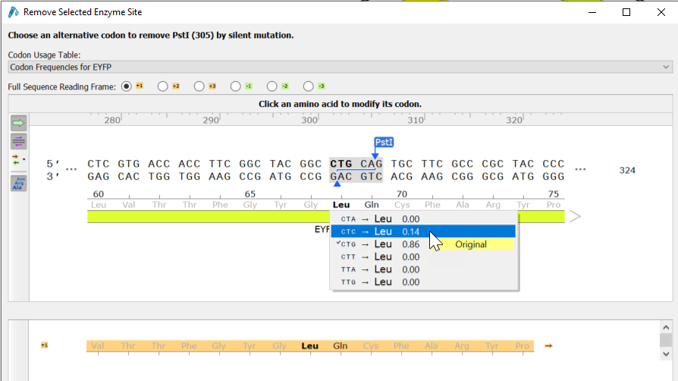
1. Addition and removal of restriction enzymes by silent mutation
The silent mutation enzyme site tool allows you to easily add or remove restriction enzyme sites from your coding sequence (add/remove restriction sites from your coding sequence without changing the translation).
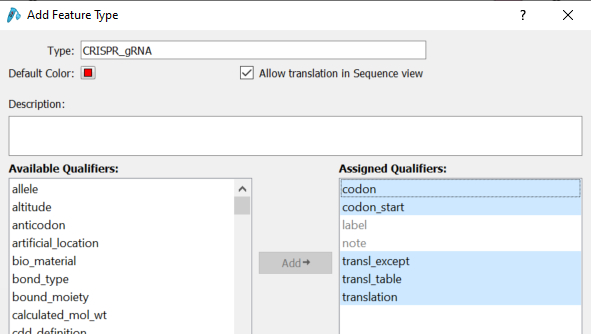
2. It is possible to define (add/delete) arbitrary feature types and modifiers.
Any feature type other than the preset feature type (GenBank) can now be handled. In addition to adding or removing feature types, you can also change the default feature color to help with identification.
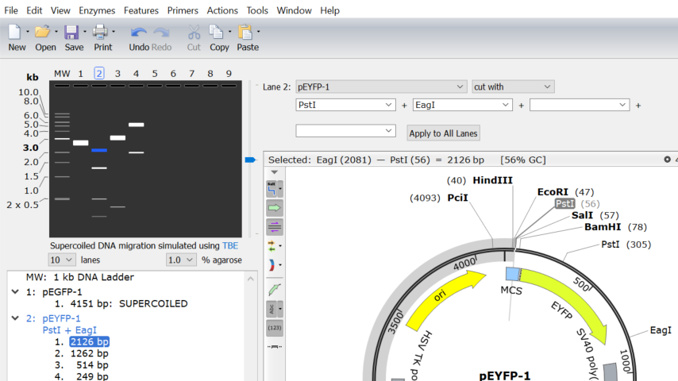
3. Agarose gel simulation can be saved as a .gel file
You can now save your agarose gel simulation as a .gel file, and you can share the gel file with other users, view it, or view and edit the gel after saving.
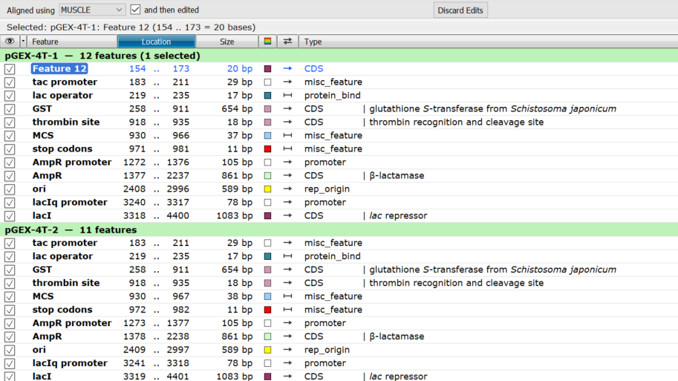
4. Functional annotation in pairwise and multiple alignments
You can view, add, and edit features for both pairwise and multiple sequence alignments. You can also switch between Alignment view and Features view to see sequences, features, and modifier information associated with each feature.
5. Enhancement of cloning simulation tools
The cloning simulation dialog has been revamped to provide more flexibility. You can add, remove, and reorder fragments within the cloning interface. Additionally, in limited cloning, Gibson Assembly, In-Fusion Cloning, and NEBuilder HiFi Assembly simulations, it is possible to set the "Orientation of Vector" to the opposite direction if necessary.
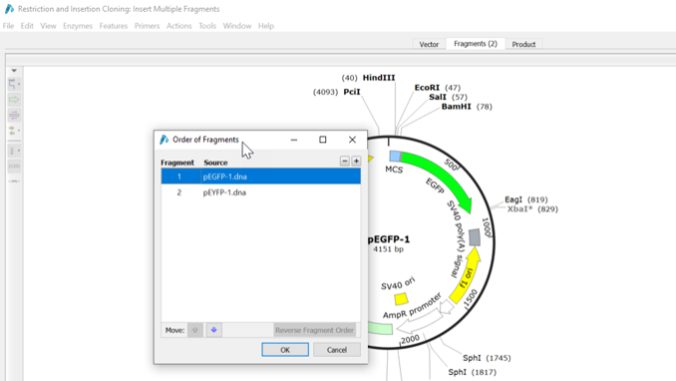
*For other functions,SnapGene6.0 release notes for more information.
|
■ Click here for product details and inquiries |



